
Edouard de Castro, Ph.D.
SIB Swiss Institute of BioInformaticsCMU, 1 Michel-Servet
Genève, GE 1211-4
Switzerland.
Fax: (+41) 22 379 5858
email: edouard.decastro@sib.swiss
publications: https://orcid.org/0000-0001-5436-7383
citations: EuropePMC author
web: Personal Wiki
LinkedIn profile
Scientist & Software Developer
Current work interests:
LLMs, AI agents,
Functional and concurrent programming, full-stack development, data visualization
Current position:
Senior software developer at Swiss Institute of BioInformatics
Currently working on:
Curation assistance & automation, Information management engines
Interest/Skill tags: AI workflows (LLMs, semantic search, RAG pipelines, agents), Functional programming, parallel computing, scala, game theory, neurosciences, biology of aging
Personal: Swiss citizen, R1b-U152 / T2b, speak English and French, born (1966) in Bruxelles (Belgium), live in Geneva (Switzerland), have 2 children (2010, 2015)
Experience / Education
Senior software developer (bioinformatics)
at the Swiss Institute of BioInformatics, Geneva, Switzerland.Swiss-Prot group of Dr. Alan Bridge.
LitSABER - curation copilot / agent - project leader and software developer (since Sept. 2024).
Wiki engines - project leader and software developer [
prime powering SwissBioPics, ViralZone, and Venomzone. hyphen for editing internal Documentation]
(since Sept. 2017).
Praise - automated curation platform - project leader and software developer
(since Apr. 2011)....
May 2002 - present
Research Scientist (molecular biology of aging)
at the University of Colorado at Boulder, USA.Laboratory of Pr. T.E. Johnson.
Oct. 1998 - April 2002
Research Scientist (molecular neurobiology)
at the Department of Biochemistry, University of Geneva, Switzerland.Laboratory of Dr. Patrick Nef.
Oct. 1997 - Sept. 1998
October 1997: Ph.D. degree in Biochemistry (with honors):
"Caenorhabditis elegans Neuronal Calcium Sensor-1: From Gene To Behavior"University of Geneva, Switzerland.
Advisors: Dr. P. Nef, Professor J. Gruenberg.
Research assistant (molecular neurobiology)
Ph.D. candidate at the Department of Biochemistry, University of Geneva, Switzerland.Advisors: Dr. P. Nef, Professor J. Gruenberg.
Jan. 1993 - Oct. 1997
Aug. 1992: Master in Biochemistry (with honors):
"Visp: 3D Molecular Visualization software"University of Geneva, Switzerland.
Advisor: Professor S. Edelstein.
Software developer / Research assistant (bioinformatics)
at the Department of Biochemistry, University of Geneva, Switzerland.Group of Professor S. Edelstein.
1992
Software developer (video game industry)
(Freelance) Sold a game to UbiSoft Entertainment (UBI).1988 - 1989
Work
Since May 2002: Software developer @ Swiss Institute of
BioInformatics
Automating and streamlining UniProtKB/Swiss-Prot protein curation workflows; Information management; data visualization:
- Some developments for UniProtKB/Swiss-Prot:
LitSABER - Using large language models (LLMs) to extract data from scientific publications (curation copilot, agent) (Scala + Java, Python).
Prime - Wiki to annotate nodes in our taxonomy / keyword hierarchies and serve portals to facilitate access to UniProtKB/Swiss-Prot. Used to edit and serve ViralZone, VenomZone and SwissBioPics (Scala + Java).
Hyphen - Wiki to edit documentation & help;
internal & for the Rhea web site (Scala + Java).
Praise - Swiss-Prot internal automated
curation platform (including biological annotation "rule" language interpretor). Used e.g. for HAMAP microbial annotation project. (Scala + Java).
publi. 21.
- Some developments for Prosite:
ScanProsite -
biological motifs detection web tool (Perl).
PSView
- domain architecture visualization; ps_scan (motif scanning) updates;
miscellaneous tools (Perl).
PSMaker - internal profile motifs building and testing platform (Perl).
publi. 9, 11,
12, 13,
14, 15,
16, 20.
- Misc:
PhiloPhylo
educational tool (in French): blast + alignment + phylogenic tree (Ruby).
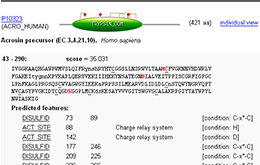
Graphical representation of PROSITE TRYPSIN_DOM hit on ACRO_HUMAN protein together with (rule based) predicted features [disulfide bridges, active sites] in ScanProsite .
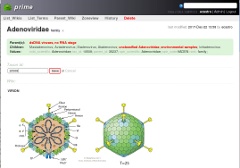
Prime web interface; modifying annotation for Adenoviridae (for ViralZone Adenoviridae page)
Oct. 1998 - April 2002: Research Scientist @ CU Boulder; Work on oxidative stress and aging in Caenorhabditis elegans. Isolation of long-lived oxidatif-stress-resistant C. elegans mutants, forward genetics... publi. 5,7, 8,10
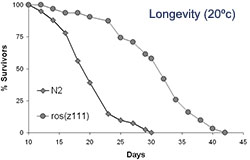
Longevity of juglone resistant C. elegans mutant ros(z111) versus wild type (N2).
1994-1998: Ph.D. + "post-doc" 1st year (research associate); Work on Caenorhabditis elegans neuronal calcium sensors. Cloning of two new C. elegans Neuronal Calcium Sensors: Ce-NCS-1 and Ce-NCS-2 publi. 2. Focusing on the study of Ce-NCS-1 publi. 4 which is highly conserved through evolution. In the C. elegans the protein, which is thought to play a role in synaptic efficacy, is expressed in some sensory neurons. Isolation of a transposon deletion derivative strain with the ncs-1 gene deleted. Analysis of the mutant that shows thermotaxis defects publi. 6.
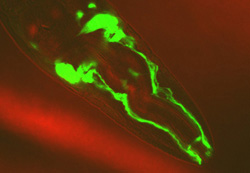
ncs-1'promoter'::GFP transgenic C. elegans (head).
1993: Ph.D. 1st year (research assistant); Work on calcium binding proteins in the nervous system. Cloning and analyzing rat NCS-1 publi. 2,4, the ortholog (100% a.a. identity) of a new calcium binding protein: chick NCS-1 (Neuronal-Calcium-Sensor 1) publi. 1, isolated in P. Nef's laboratory.
1992: Master in Biochemistry;
Work on molecular computer graphics. Creation of
ViSP (Visualization System
for Proteins) a software for 3D visualization of
macromolecules (written in C on a SGI Irix 4D/35
computer).
This program was used to make 3D structures illustrations,
for example in:
-
NMR Structure of a Specific DNA Complex of Zn-Containing
DNA Binding Domain of oinformatics
. 2020 Mar 1;36(6):1896-1901.
doi: 10.1093/bioinformatics/btz817.GATA-1.
J.G. Omichinski et al, Science 261: 438 (1993).
-
Structures of larger proteins, protein-ligand and
protein-DNA complexes by multidimensional heteronuclear
NMR.
G.M. Clore et al, Protein Science 3: 372 (1994).
- Cover of Guidebook to the Calcium Binding Proteins. Edited by M.R. Cello, Oxford Univeristy Press, 1996.
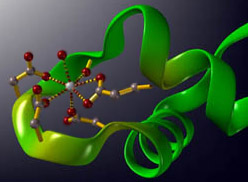
Image created using ViSP (Ca2+ binding on a EF-Hand structure in the protein parvalbumin)
1988-89: Game developer (freelance). Development and design of Othello Killer [Othello/Reversi strategic board game program where users can play against the computer (level: intermediate-advanced)] written in C for the AMIGA computer, published in 1989 by UbiSoft Entertainment.
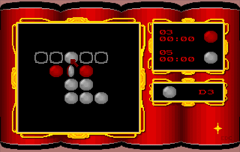
"Othello Killer" game screenshot
1989: Development of Digital Studio (not commercialized), digital multitrack recorder software used to digitize sounds and music for Othello Killer, using a custom made audio analog to digital converter made by Curzio Bossi (BTZ electronic, Geneva, Switzerland).
Skills
Software development:
- Programming languages: Scala, Node.js / JavaScript, Perl, Python, Java, C/C++, Ruby.- Programming Technologies/frameworks/tools/Paradigms: Functional programming, Object-oriented programming, Futures parallelization, AI via OpenAI API: chat completions, embeddings, fine-tuning, custom AI agent creation (integrating LLMs, semantic search, RAG, biological APIs, and schedulers), HTML / CSS + JavaScript (w/ fetch API, Promises, web components, templates), google charts, Java Virtual Machine, REST, http, Relational Databases [PostgreSQL, Oracle], Full-Stack Development, sparql, rdf, OpenGL, jdbc, Automated Testing [ScalaTest~WordSpec], Regexps, ant, sbt, (HTML5) Canvas 2D, Jira, Confluence, git, Jenkins, ChatGPT "code interpretor".
- IDEs: IntelliJ IDEA (for Scala, Java, Perl), Xcode
- OSes: Linux, macos, iOS.
- Software: Adobe Photoshop, Illustrator, BioInformatics tools (blast, profiles / Hidden Markov models etc).
'Likes: Code with tests!, minimal mutability, minimal "framework-ness"; explicitness; valid html.
Miscellaneous:
Genetics, Molecular Biology, Neurobiology, Biology of ageing, Biochemistry, Caenorhabditis elegans model organism, Proteomics, Combinatorial game theory (decision trees, minimax, alpha-beta pruning), Monte Carlo simulation.Fellowships / Grants
September 1999-, 1.5 years. Swiss National Science
Foundation "Advanced researcher fellowship"
"Identification of genes involved in oxidative
stress response and aging in Caenorhabditis elegans"
, fellowship n°823A-056586.
September 1998-, 1 year. Swiss National Science
Foundation "Beginner researcher fellowship"
"Characterization and analysis of differentially
expressed genes in age-1 long-lived Caenorhabditis elegans
mutants", fellowship n°81GE-51690.
January 1997-, 8 months. Roche Research Foundation
Fellowship with Sandoz Stiftung participation
"Functional analysis of the neuronal calcium
sensor-1 with a Caenorhabditis elegans knock-out
mutant."
Publications
36. Edouard de Castro; Chantal Hulo; Patrick Masson; Andrea Auchincloss; Alan Bridge; Philippe Le Mercier. ViralZone 2024 provides higher-resolution images and advanced virus-specific resources. Nucleic Acids Research, 2023, gkad946, https://doi.org/10.1093/nar/gkad946
35. Elisabeth Coudert; Sebastien Gehant; Edouard de Castro; Monica Pozzato; Delphine Baratin; Teresa Batista Neto; Christian J.A. Sigrist; Nicole Redaschi; Alan Bridge; The UniProt Consortium. Annotation of biologically relevant ligands in UniProtKB using ChEBI Bioinformatics, Volume 39, Issue 1, January 2023, btac793, https://doi.org/10.1093/bioinformatics/btac793
34. Philippe Le Mercier, Jerven Bolleman, Edouard de Castro, Elisabeth Gasteiger, Parit Bansal, Andrea H Auchincloss, Emmanuel Boutet, Lionel Breuza, Cristina Casals-Casas, Anne Estreicher, Marc Feuermann, Damien Lieberherr, Catherine Rivoire, Ivo Pedruzzi, Nicole Redaschi, Alan Bridge SwissBioPics—an interactive library of cell images for the visualization of subcellular location data Database, Volume 2022, 2022, baac026, doi: 10.1093/database/baac026
33. Feuermann M, Boutet E, Morgat A, Axelsen K, Bansal P. Bolleman J, de Castro E, Coudert E, Gasteiger E, Gehant S, Lieberherr D, Lombardot T, Neto T, Pedruzzi S, Poux S, Pozzato M, Redaschi N, Alan Bridge; UniProt Consortium. Diverse Taxonomies for Diverse Chemistries: Enhanced Representation of Natural Product Metabolism in UniProtKB Metabolites 2021, 11(1), 48. doi: 10.3390/metabo11010048
32.
Le Mercier P, Hulo C, Masson P, de Castro E.
ViralZone : le numérique au service du partage des savoirs
Virologie 2020; xx(x) : 1-4 doi:10.1684/vir.2020.0873
31. Morgat A, Lombardot T, Coudert E, Axelsen K, Neto TB, Gehant S, Bansal P, Bolleman J, Gasteiger E, de Castro E, Baratin D, Pozzato M, Xenarios I, Poux S, Redaschi N, Bridge A; UniProt Consortium. Enzyme annotation in UniProtKB using Rhea Bioinformatics. 2020 Mar 1;36(6):1896-1901. doi: 10.1093/bioinformatics/btz817.
30. Bolleman J, de Castro E, Baratin D, Gehant S, Cuche BA, Auchincloss AH, Coudert E, Hulo C, Masson P, Pedruzzi I, Rivoire C, Xenarios I, Redaschi N, Bridge A. HAMAP as SPARQL rules-A portable annotation pipeline for genomes and proteomes Gigascience. 2020 Feb 1;9(2):giaa003. doi: 10.1093/gigascience/giaa003.
29. Morgat A, Lombardot T, Coudert E, Axelsen K, Neto TB, Gehant S, Bansal P, Bolleman J, Gasteiger E, de Castro E, Baratin D, Pozzato M, Xenarios I, Poux S, Redaschi N, Bridge A; UniProt Consortium. Enzyme annotation in UniProtKB using Rhea Bioinformatics. 2019 Nov 5. pii: btz817. doi: 10.1093/bioinformatics/btz817.
28. Hulo C, Masson P, Toussaint A, Osumi-Sutherland D, de Castro E, Auchincloss AH, Poux S, Bougueleret L, Xenarios I, Le Mercier P. Bacterial Virus Ontology; Coordinating across Databases Viruses. 2017 May 23;9(6). pii: E126. doi: 10.3390/v9060126.
27. Hulo C, Masson P, de Castro E, Auchincloss AH, Foulger R, Poux S, Lomax J, Bougueleret L, Xenarios I, Le Mercier P. The ins and outs of eukaryotic viruses: Knowledge base and ontology of a viral infection PLoS One. 2017 Feb 16;12(2):e0171746. doi: 10.1371/journal.pone.0171746. eCollection 2017.
26. The SIB Swiss Institute of Bioinformatics' resources: focus on curated databases Nucleic Acids Res. 2016 Jan 4;44(D1):D27-37. doi: 10.1093/nar/gkv1310. Epub 2015 Nov 28.
25. Pedruzzi I, Rivoire C, Auchincloss AH, Coudert E, Keller G, de Castro E, Baratin D, Cuche BA, Bougueleret L, Poux S, Redaschi N, Xenarios I, Bridge A. HAMAP in 2015: updates to the protein family classification and annotation system Nucleic Acids Res. 2015 Jan;43(Database issue):D1064-70 [10.1093/nar/gku1002] Epub 2014 Oct 27 (2014).
24. Masson P, Hulo C, de Castro E, Foulger R, Poux S, Bridge A, Lomax J, Bougueleret L, Xenarios I, Le Mercier P. An integrated ontology resource to explore and study host-virus relationships PLoS One. 2014 Sep 18;9(9):e108075 [10.1371/journal.pone.0108075] eCollection (2014).
23. Cannarozzi G, Plaza-Wuethrich S, Esfeld K, Larti S, Wilson YS, Girma D, de Castro E, Chanyalew S, Bloesch R, Farinelli L, Lyons E, Schneider M, Falquet L, Kuhlemeier C, Assefa K, Tadele Z. Genome and transcriptome sequencing identifies breeding targets in the orphan crop tef (Eragrostis tef) BMC Genomics. 2014 Jul 9;15:581 [10.1186/1471-2164-15-581] (2014).
22. Masson P, Hulo C, De Castro E, Bitter H, Gruenbaum L, Essioux L, Bougueleret L, Xenarios I, Le Mercier P. ViralZone: recent updates to the virus knowledge resource Nucleic Acids Res. 2013 Jan;41(Database issue):D579-83 [10.1093/nar/gks1220] (2013).
21. Pedruzzi I, Rivoire C, Auchincloss AH, Coudert E, Keller G, de Castro E, Baratin D, Cuche BA, Bougueleret L, Poux S, Redaschi N, Xenarios I, Bridge A; UniProt Consortium. HAMAP in 2013, new developments in the protein family classification and annotation system Nucleic Acids Res. 2013 Jan;41(Database issue):D584-9 [10.1093/nar/gks1157] (2013).
20. Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. New and continuing developments at PROSITE Nucleic Acids Res. 2013 Jan;41(Database issue):D344-7 [doi: 10.1093/nar/gks1067] (2013).
19. Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H. ExPASy: SIB bioinformatics resource portal Nucleic Acids Res. 2012 Jul;40(Web Server issue):W597-603 [10.1093/nar/gks400] (2012).
18. Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, ... Yong SY. InterPro in 2011: new developments in the family and domain prediction database Nucleic Acids Res. 2012 Jan;40(Database issue):D306-12 [10.1093/nar/gkr948] (2011).
17. Chantal Hulo, Edouard de Castro, Patrick Masson, Lydie Bougueleret, Amos Bairoch, Ioannis Xenarios, Philippe Le Mercier ViralZone: a knowledge resource to understand virus diversity Nucleic Acids Res. 39(suppl 1): D576-D582 [10.1093/nar/gkq901] (2010).
16. Sigrist CJA, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 38(Database issue): D161-6 [10.1093/nar/gkp885] (2010).
15. Lima T., Auchincloss A.H., Coudert E., Keller G., Michoud K., Rivoire C., Bulliard V., de Castro E., Lachaize C., Baratin D., Phan I., Bougueleret L., Bairoch A. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 37: D471-D478 [10.1093/nar/gkn661] (2009).
14. Nicolas Hulo, Amos Bairoch, Virginie Bulliard, Lorenzo Cerutti, Beatrice A. Cuche, Edouard de Castro, Corinne Lachaize, Petra S. Langendijk-Genevaux, and Christian J. A. Sigrist. The 20 years of PROSITE Nucleic Acids Res. 36(Database issue): D245-D249 [doi:10.1093/nar/gkm977] (January 2008).
13. Edouard de Castro, Christian J.A. Sigrist, Alexandre Gattiker, Virginie Bulliard, Petra S. Langendijk-Genevaux, Elisabeth Gasteiger, Amos Bairoch, Nicolas Hulo. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34(Web Server issue):W362-W365 [doi:10.1093/nar/gkl124] (July 2006).
12. Hulo N, Bairoch A, Bulliard V, Cerutti L, de Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJ. The PROSITE database. Nucleic Acids Res. 34(Database issue): D227-30 [10.1093/nar/gkj063] (Jan 2006).
11. Sigrist C.J.A., de Castro E., Langendijk-Genevaux P.S., Le Saux V., Bairoch A., Hulo N. ProRule: a new database containing functional and structural information on PROSITE profiles. Bioinformatics. 21(21): 4060-6 (2005, Nov. 1).
10. Edouard de Castro, Sarah Hegi de Castro, Thomas E. Johnson. Isolation of long-lived mutants in Caenorhabditis elegans using juglone resistance selection. Free Radical Biology and Medicine 37(2): 139-145 (2004).
9. Nicolas Hulo, Christian J. A. Sigrist, Virginie Le Saux, Petra S. Langendijk-Genevaux, Lorenza Bordoli1, Alexandre Gattiker, Edouard De Castro, Philipp Bucher, Amos Bairoch. Recent improvements to the PROSITE database. Nucleic Acids Research 32: D134-D137 (2004).
8. Thomas E. Johnson, Sam Henderso, Shin Murakami, Edouard de Castro, Sarah Hegi de Castro, James Cypser, Brad Rikke, Pat Tedesco, Chris Link. Longevity genes in the nematode caenorhabditis elegans also mediate increased resistance to stress and prevent disease [review] J. of Inherited Metabolic Disease 25: 197-206 (2002).
7. Thomas E. Johnson, Edouard de Castro, Sarah Hegi de Castro, James Cypser, Sam Henderson, and Pat Tedesco. Relationship between increased longevity and stress resistance as assessed through gerontogene mutations in Caenorhabditis elegans. [review] Experimental Gerontology 36: 1609-1617 (2001).
6. Marie Gomez* & Edouard De Castro*, Ernesto Guarin, Hiroyuki Sasakura, Atsushi Kuhara, Ikue Mori, Tamas Bartfai, Cornelia I. Bargmann, Patrick Nef. Ca2+-Signaling via the Neuronal Calcium Sensor-1 Regulates Associative Learning and Memory in C. elegans. Neuron 30 (1): 241-248 (April 2001). *These authors contributed equally to this work.
5. T.E. Johnson, J. Cypser, E. de Castro, S. de Castro, S. Henderson, S. Murakami, B. Rikke, P. Tedesco, C. Link. Gerontogenes mediate health and longevity in nematodes through increasing resistance to environmental toxins and stressors. [review] Experimental Gerontology 35: 687-694 (2000).
Edouard de Castro. Caenorhabditis elegans Neuronal Calcium Sensor-1: From Gene to Behavior. Ph.D. Thesis (n° 2943). Université de Genève (1997).
4. Schaad N, De Castro E, Nef S, Hegi S, Hinrichsen R, Martone M, Ellisman M, Sikkink R, Rusnak F, Sygush J, Nef P. Direct modulation of calmodulin targets by the neuronal calcium sensor NCS-1. Proc. Natl. Acad. Sci. USA 93: 9253-9258 (1996).
3. Nef S, Allaman I, Fiumelli H, De Castro E, Nef P. Olfaction in birds: differential embryonic expression of nine putative odorant receptor genes in the avian olfactory system. Mechanisms of Development 55: 65-77 (1996).
2. De Castro E, Nef S, Fiumelli H, Lenz SE, Kawamura S, Nef P. Regulation of rhodopsin phosphorylation by a family of neuronal calcium sensors. Biochem. Biophys. Res. Commun. 216: 133-140 (1995).
1. Nef S, Fiumelli H, De Castro E, Raes M-B, Nef P. Identification of a neuronal calcium sensor (NCS-1) possibly involved in the regulation of receptor phosphorylation. J. Recept. Res. 15(1-4): 365-378 (1995).